import pandas as pd
import numpy as np
import lightgbm as lgb
from polynomial_boosting import PolynomialBoostingModel
import matplotlib.pyplot as plt
from sklearn.ensemble import GradientBoostingRegressor
from sklearn.datasets import fetch_california_housing
from sklearn.model_selection import train_test_splitGradient Boosting with Polynomial Decision Trees
Fit an arbitrarily complex Polynomial in Decision Tree leaf nodes, rather than a flat line. Code for polynomial_boosting can be found here.
np.random.seed(123)
def f(X):
return X**2
N = 1000
X = np.random.uniform(size=(N,1)) * 6 - 3
#model requires that `y` is a matrix and not a vector (contrary to sklearn models)
y = f(X) + np.random.normal(size=(N,1)) * 0.5
line = np.linspace(-3,3,250)
plt.scatter(X[:,0],y[:,0],s=1)
plt.plot(line,f(line),c="red")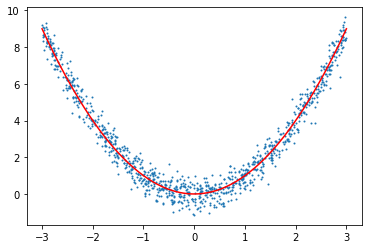
model = PolynomialBoostingModel(1, #polynomial_level
0.25, #learning rate
5.0, #regression regularization
100, #n_trees
10, #min_samples_leaf
1.0, #goss alpha
0.0, #goss beta
1) #random seed
model.fit_fast(X,y)
preds = model.predict_fast(line.reshape(-1,1)).reshape(-1)
plt.scatter(X[:,0],y[:,0],s=0.05)
plt.plot(line,f(line),c="red")
plt.plot(line,preds,c="green")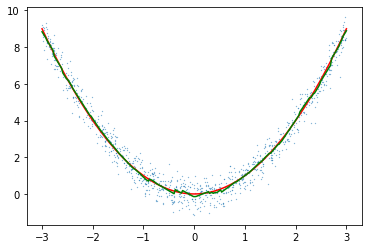
model2 = GradientBoostingRegressor(max_depth=1, n_estimators=100, learning_rate = 0.25)
model2.fit(X,y.reshape(-1))
preds2 = model2.predict(line.reshape(-1,1)).reshape(-1)
plt.scatter(X[:,0],y[:,0],s=0.05)
plt.plot(line,f(line),c="red")
plt.plot(line,preds2,c="green")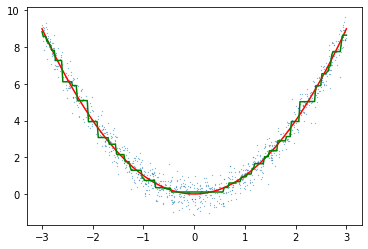
lgb_dataset = lgb.Dataset(X, label=y.reshape(-1), params={'linear_tree': True})
lgb_params = {
"objective": "regression",
"metric": "l2",
"num_iterations": 100,
"num_leaves": 2,
"learning_rate": 0.25,
"linear_lambda": 5.0,
"min_data_in_leaf": 10,
"verbosity": -1
}
model3 = lgb.train(lgb_params, lgb_dataset)
preds3 = model3.predict(line.reshape(-1,1)).reshape(-1)
plt.scatter(X[:,0],y[:,0],s=0.05)
plt.plot(line,f(line),c="red")
plt.plot(line,preds3,c="green")/Library/Frameworks/Python.framework/Versions/3.10/lib/python3.10/site-packages/lightgbm/engine.py:172: UserWarning: Found `num_iterations` in params. Will use it instead of argument
_log_warning(f"Found `{alias}` in params. Will use it instead of argument")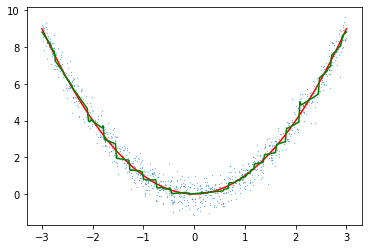
fig, axes = plt.subplots(nrows=1, ncols=3, figsize=(12, 6))
axes[0].scatter(X[:,0], y[:,0], color='blue', label='Training Data', s=0.1)
axes[0].plot(line, f(line), color='green', label='True Function')
axes[0].plot(line, preds, color='red', label='Predicted')
axes[0].set_title('Polynomial (linear) Boosting - MSE: {:.4f}'.format(np.mean((preds-f(line))**2)))
axes[0].set_xlabel('X')
axes[0].set_ylabel('Y')
axes[0].legend()
axes[1].scatter(X[:,0], y[:,0], color='blue', label='Training Data', s=0.1)
axes[1].plot(line, f(line), color='green', label='True Function')
axes[1].plot(line, preds2, color='red', label='Predicted')
axes[1].set_title('Sklearn Gradient Boosting - MSE: {:.4f}'.format(np.mean((preds2-f(line))**2)))
axes[1].set_xlabel('X')
axes[1].set_ylabel('Y')
axes[1].legend()
axes[2].scatter(X[:,0], y[:,0], color='blue', label='Training Data', s=0.1)
axes[2].plot(line, f(line), color='green', label='True Function')
axes[2].plot(line, preds3, color='red', label='Predicted')
axes[2].set_title('LGBM Linear Boosting - MSE: {:.4f}'.format(np.mean((preds3-f(line))**2)))
axes[2].set_xlabel('X')
axes[2].set_ylabel('Y')
axes[2].legend()
plt.tight_layout()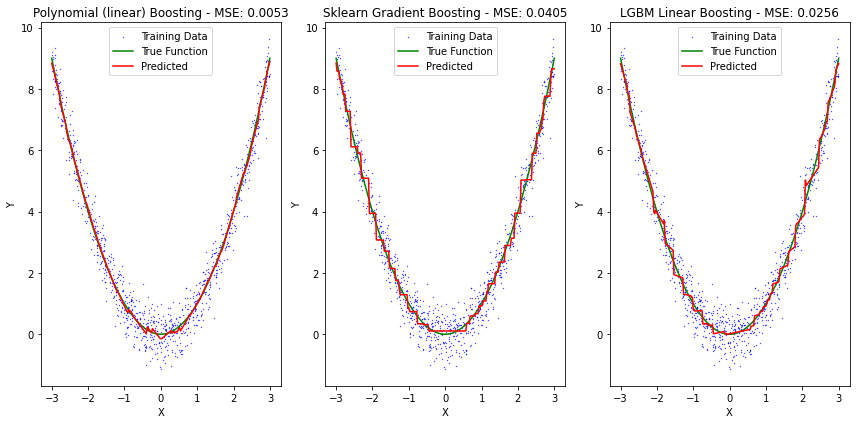
Xd, yd = fetch_california_housing(return_X_y=True)
#small train set, as the model is still quite slow
X_train, X_test, y_train, y_test = train_test_split(Xd, yd, test_size=0.975, random_state=123)
Xm = np.mean(X_train,0).reshape(1,-1)
Xs = np.std(X_train,0).reshape(1,-1)
ym = np.mean(y_train)
ys = np.std(y_train)
X_train = (X_train - Xm) / Xs
y_train = (y_train - ym) / ys
X_test = (X_test - Xm) / Xsd_model = PolynomialBoostingModel(1, #polynomial level
0.25, #learning rate
5.0, #regression regularization
100, #n_trees
10, #min_samples_leaf
1.0, #goss alpha
0.0, #goss beta
1) #random seed
d_model.fit_fast(X_train,y_train.reshape(-1,1))
d_preds = d_model.predict_fast(X_test).reshape(-1) * ys + ymd_model2 = GradientBoostingRegressor(max_depth=1, n_estimators=100, learning_rate = 0.25)
d_model2.fit(X_train,y_train)
d_preds2 = d_model2.predict(X_test) * ys + ymlgb_dataset = lgb.Dataset(X_train, label=y_train.reshape(-1), params={'linear_tree': True})
lgb_params = {
"objective": "regression",
"metric": "l2",
"num_iterations": 100,
"num_leaves": 2,
"learning_rate": 0.25,
"linear_lambda": 5.0,
"min_data_in_leaf": 10,
"verbosity": -1
}
d_model3 = lgb.train(lgb_params, lgb_dataset)
d_preds3 = d_model3.predict(X_test).reshape(-1) * ys + ym/Library/Frameworks/Python.framework/Versions/3.10/lib/python3.10/site-packages/lightgbm/engine.py:172: UserWarning: Found `num_iterations` in params. Will use it instead of argument
_log_warning(f"Found `{alias}` in params. Will use it instead of argument")np.mean((d_preds-y_test)**2)0.40638200743172254np.mean((d_preds2-y_test)**2)0.4231840786265534np.mean((d_preds3-y_test)**2) 0.41076314048606744feature_names = fetch_california_housing()["feature_names"]
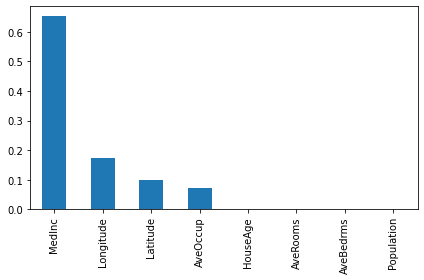
importances = pd.Series(d_model.get_feature_importances()[:8], index = feature_names)
importances.sort_values(ascending=False).plot(kind="bar")
plt.tight_layout()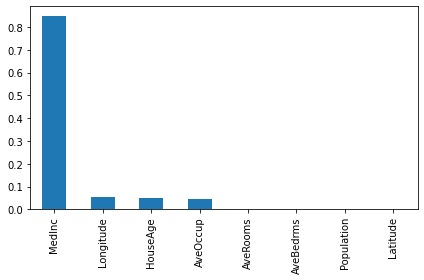
importances2 = pd.Series(d_model2.feature_importances_, index = feature_names)
importances2.sort_values(ascending=False).plot(kind="bar")
plt.tight_layout()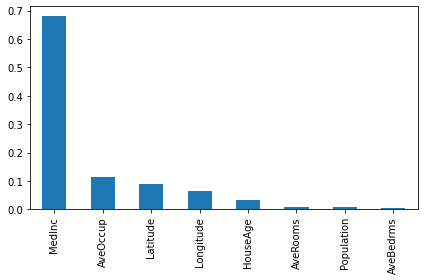
importances3 = pd.Series(d_model3.feature_importance()/np.sum(d_model3.feature_importance()), index=feature_names)
importances3.sort_values(ascending=False).plot(kind="bar")
plt.tight_layout()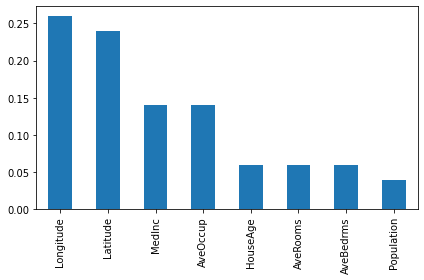
Larger dataset
Xd, yd = fetch_california_housing(return_X_y=True)
#small train set, as the model is still quite slow
X_train, X_test, y_train, y_test = train_test_split(Xd, yd, test_size=0.2, random_state=123)
Xm = np.mean(X_train,0).reshape(1,-1)
Xs = np.std(X_train,0).reshape(1,-1)
ym = np.mean(y_train)
ys = np.std(y_train)
X_train = (X_train - Xm) / Xs
y_train = (y_train - ym) / ys
X_test = (X_test - Xm) / XsUse GOSS sampling to keep computational demand feasible (see the LightGBM paper, algorithm 2)
Currently, not using specific weights for the randomly sampled subset in GOSS appears to outperform the exact GOSS proposal. This might be due to the regression models not being fit with those weights as well but only the total node losses.
d_model = PolynomialBoostingModel(
1, #polynomial level
0.25, #learning rate
5.0, #regression regularization
100, #n_trees
10, #min_samples_leaf
0.2, #goss alpha
0.025, #goss beta
1) #random seed
d_model.fit_fast(X_train,y_train.reshape(-1,1))
d_preds = d_model.predict_fast(X_test).reshape(-1) * ys + ymd_model2 = GradientBoostingRegressor(max_depth=1, n_estimators=100, learning_rate = 0.25, min_samples_leaf=10)
d_model2.fit(X_train,y_train)
d_preds2 = d_model2.predict(X_test) * ys + ymlgb_dataset = lgb.Dataset(X_train, label=y_train.reshape(-1), params={'linear_tree': True})
lgb_params = {
"objective": "regression",
"metric": "l2",
"num_iterations": 100,
"num_leaves": 2,
"learning_rate": 0.25,
"linear_lambda": 5.0,
"data_sampling_strategy": "goss",
"top_rate": 0.2, #=goss alpha
"other_rate": 0.025, #=goss beta
"verbosity": -1
}
d_model3 = lgb.train(lgb_params, lgb_dataset)
d_preds3 = d_model3.predict(X_test).reshape(-1) * ys + ym/Library/Frameworks/Python.framework/Versions/3.10/lib/python3.10/site-packages/lightgbm/engine.py:172: UserWarning: Found `num_iterations` in params. Will use it instead of argument
_log_warning(f"Found `{alias}` in params. Will use it instead of argument")d_model4 = PolynomialBoostingModel(
2, #polynomial level
0.25, #learning rate
5.0, #regression regularization
100, #n_trees
10, #min_samples_leaf
0.2, #goss alpha
0.025, #goss beta
1) #random seed
d_model4.fit_fast(X_train,y_train.reshape(-1,1))
d_preds4 = d_model4.predict_fast(X_test).reshape(-1) * ys + ymnp.mean((d_preds-y_test)**2) #boosted linear trees (p=1)0.34790501763806014np.mean((d_preds2-y_test)**2) #sklearn boosted trees (p=0)0.3797511677878719np.mean((d_preds3-y_test)**2) #lgbm boosted trees (p=1)0.3640894459649956np.mean((d_preds4-y_test)**2) #boosted quadratic trees (p=2)0.33865369672689777feature_names = fetch_california_housing()["feature_names"]
importances = pd.Series(d_model.get_feature_importances()[:8], index = feature_names)
importances.sort_values(ascending=False).plot(kind="bar")
plt.tight_layout()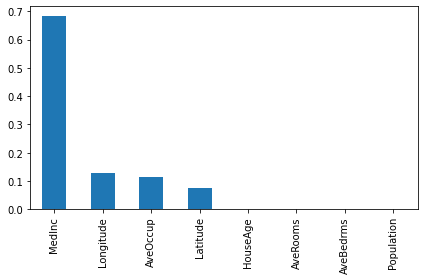
importances2 = pd.Series(d_model2.feature_importances_, index = feature_names)
importances2.sort_values(ascending=False).plot(kind="bar")
plt.tight_layout()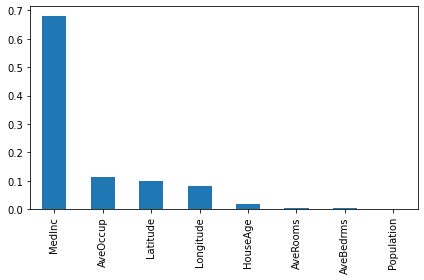
importances3 = pd.Series(d_model3.feature_importance()/np.sum(d_model3.feature_importance()), index=feature_names)
importances3.sort_values(ascending=False).plot(kind="bar")
plt.tight_layout()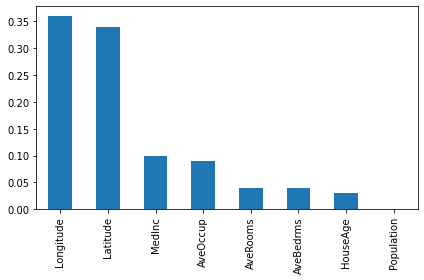
importances4 = pd.Series(d_model4.get_feature_importances()[:8], index = feature_names)
importances4.sort_values(ascending=False).plot(kind="bar")
plt.tight_layout()